Dexamethasone Benchmark Code Notebooks
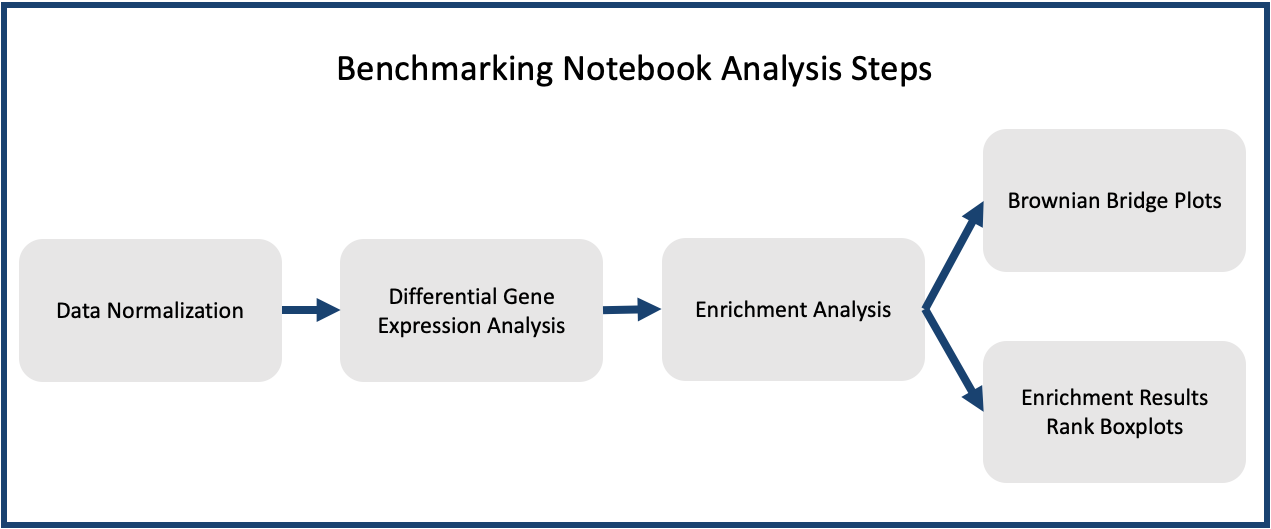
All code used to benchmark data can be found on this page in the form of Jupyter Notebooks (.ipynb files) roughly following the pipeline steps shown in the figure to the right.
To run a given notebook, you can either clone this Github repository in its entirety, or download only the notebook and relevant files you need. For the L1000 notebooks, all data is hosted in an Amazon S3 bucket, and the notebook will automatically download the required data the first time the notebook is run. Each subsequent run will use the downloaded data.
Pre-computed example HTML files are also available for each notebook, and showcase both the code and the expected tabular and visual outputs.
Retrieving L1000 Data
All L1000 profiles used in the benchmarking notebooks can be downloaded directly from an Amazon S3 bucket. The download links are provided in the SigCom LINCS Metadata, which can be accessed from the SigCom LINCS API.
The following notebook provides a guide for downloading L1000 chemical, overexpression, shRNA knockdown, and CRISPR knockout profiles.
- Retrieving L1000 Data via SigCom LINCS API Notebook Download and Example HTML
Differential Gene Expression Analysis Methods
RNA-seq Data
- Benchmarking Dexamethasone Notebook Download and Example HTML
L1000 Data
Metadata for all relevant L1000 data profiles and signatures can be found on the Data page.
-
Benchmarking DEG Methods for L1000 CRISPR KO Data
- NR0B1 Notebook Download and Example HTML
- NR1I2 Notebook Download and Example HTML
-
Benchmarking DEG Methods for L1000 Chemical Perturbation Data
- Dexamethasone Notebook Download and Example HTML
L1000 Consensus Signatures
- Benchmarking L1000 consensus signatures Notebook Download and Example HTML