About the Dexamethasone Benchmark (Dex-Benchmark) Resource
The volume and diversity of transcriptomics datasets is rapidly growing. At the same time, hundreds of new tools are published each year to perform various steps in transcriptomics data analysis, such as identifying differentially expressed genes, performing gene set enrichment analysis, and visualizing transcriptomics data as heatmaps and scatter plots. In order to evaluate methods that perform these tasks, there is a need for developing objective benchmarks that can be used to compare the quality of tools and algorithms to maximally and accurately extract biological knowledge from these data.
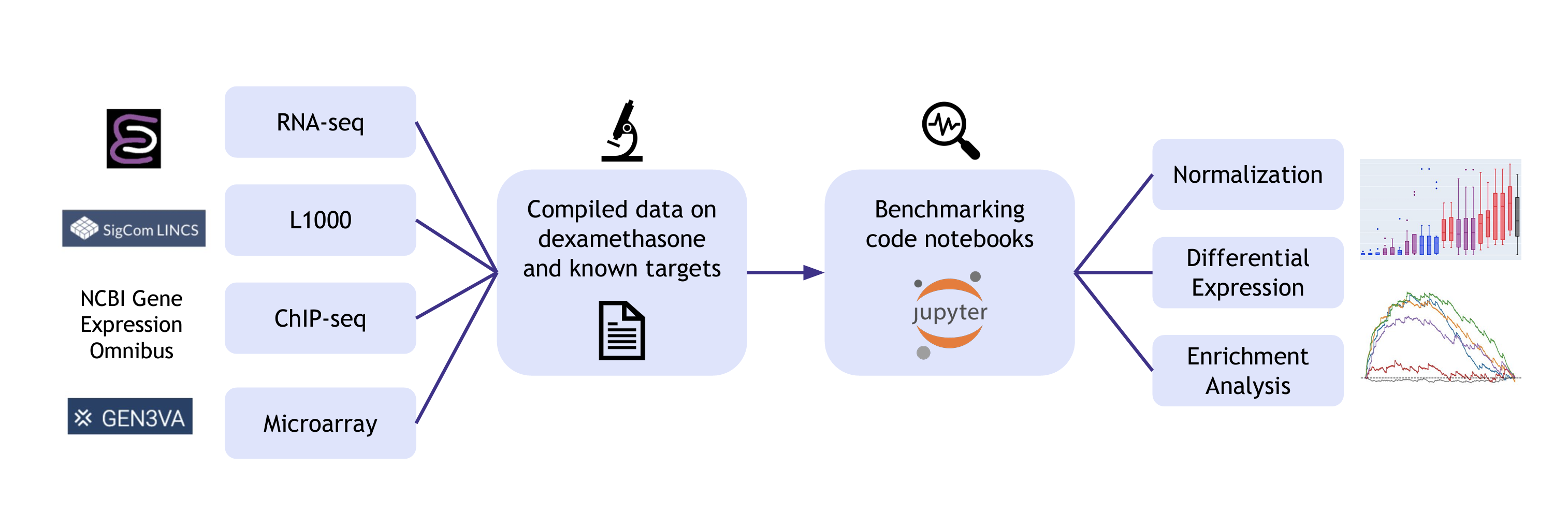
The goal of this resource is to provide the community with datasets and template workflows needed for benchmarking different tools for gene expression analysis. Gene expression and transcription factor binding data for the drug dexamethasone and its known target, the Glucocorticoid Receptor (GR) (NR3C1), are provided on the landing page along with code that demonstrates how to perform benchmarks for comparing gene expression analysis steps, such as alignment, data normalization, unsupervised cluster identification, differential expression analysis, and enrichment analysis. The sources for the curated datasets include SigCom LINCS1, ENCODE2,3, and the Gene Expression Omnibus4 (GEO) with processed data from CREEDS5, GEN3VA6, and ChEA7. Some of the provided data is used for the template workflows that compare state-of-the-art algorithms currently used by the community to perform transcriptomics analyses. Users can also benchmark additional methods by customizing the Jupyter Notebooks for their own needs. Overall, the Dex-Benchmark Resource provides a step forward toward improving the quality of bioinformatics workflow for the betterment of knowledge extraction from omics datasets.
Read the Paper
Xie Z, Chen C, Ma'ayan A. Dex-Benchmark: datasets and code to evaluate algorithms for transcriptomics data analysis. PeerJ. 2023 Nov 8;11:e16351. doi: 10.7717/peerj.16351. PMID: 37953774; PMCID: PMC10638921.
References
[1] Evangelista JE, Clarke DJB, Xie Z, Lachmann A, Jeon M, Chen K, Jagodnik KM, Jenkins SL, Kuleshov MV, Wojciechowicz ML, Schürer SC, Medvedovic M, Ma’ayan A. SigCom LINCS: data and metadata search engine for a million gene expression signatures. Nucleic Acids Research, 2022; gkac328, https://doi.org/10.1093/nar/gkac328
[2] ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):57-74. doi: 10.1038/nature11247. PMID: 22955616; PMCID: PMC3439153.
[3] Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, Baymuradov UK, Graham K, Litton C, Miyasato SR, Strattan JS, Jolanki O, Lee JW, Tanaka FY, Adenekan P, O'Neill E, Cherry JM. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020 Jan 8;48(D1):D882-D889. doi: 10.1093/nar/gkz1062. PMID: 31713622; PMCID: PMC7061942.
[4] Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5.
[5] Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz M, Utti V, Jagodnik K, Kropiwnicki E, Wang Z, Ma'ayan A (2019) ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Research. doi: 10.1093/nar/gkz446
[6] Wang, Z., Monteiro, C. D., Jagodnik, K. M., Fernandez, N. F., Gundersen, G. W., ... & Ma'ayan, A. (2016) Extraction and Analysis of Signatures from the Gene Expression Omnibus by the Crowd. Nature Communications doi: 10.1038/ncomms12846
[7] Gundersen GW, Jagodnik KM, Woodland H, Fernandez NF, Sani K, Dohlman AB, Ung PM, Monteiro CD, Schlessinger A, Ma'ayan A. GEN3VA: aggregation and analysis of gene expression signatures from related studies. BMC Bioinformatics. 2016 Nov 15;17(1):461. doi: 10.1186/s12859-016-1321-1. PMID: 27846806; PMCID: PMC5111283.