Omics Mining for the Illuminating the Druggable Genome Project
Target Functional Predictions
Methods: To predict functions for the under-studied kinases, GPCRs and ion channels we used a co-expression network created from RNA-seq data processed from GEO/SRA. An example is provided to show how biological processes from the gene ontology (GO) can be predicted. First, we processed all human and mouse RNA-seq samples currently (2/2017) available from GEO/SRA. Next, we calculated all pairwise gene correlations from the all the processed RNA-seq expression data. We then iterate over 5192 GO biological process terms where for each gene we calculated the average correlation with the genes in the gene set. The values are stored in a matrix C for all i genes and j biological processes. We then z-score normalized all the columns of C. Finally, for each gene in C we ranked terms based on their gene population z-score. Terms with high z-scores are predicted to be true annotations for the gene.
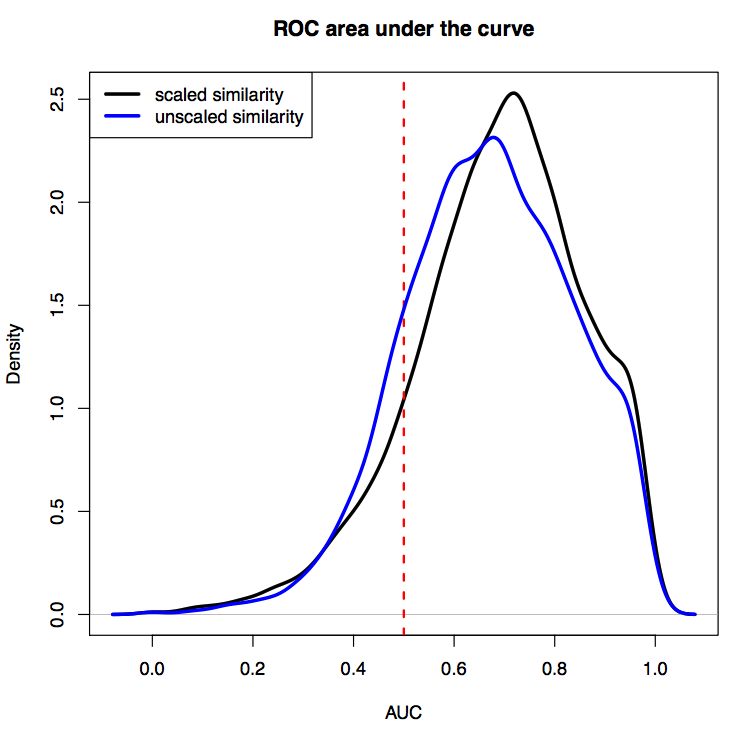
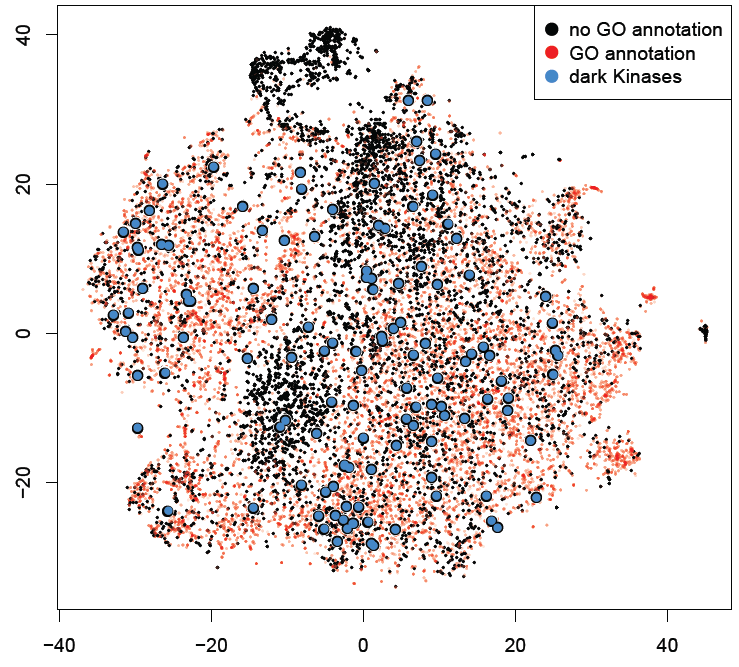
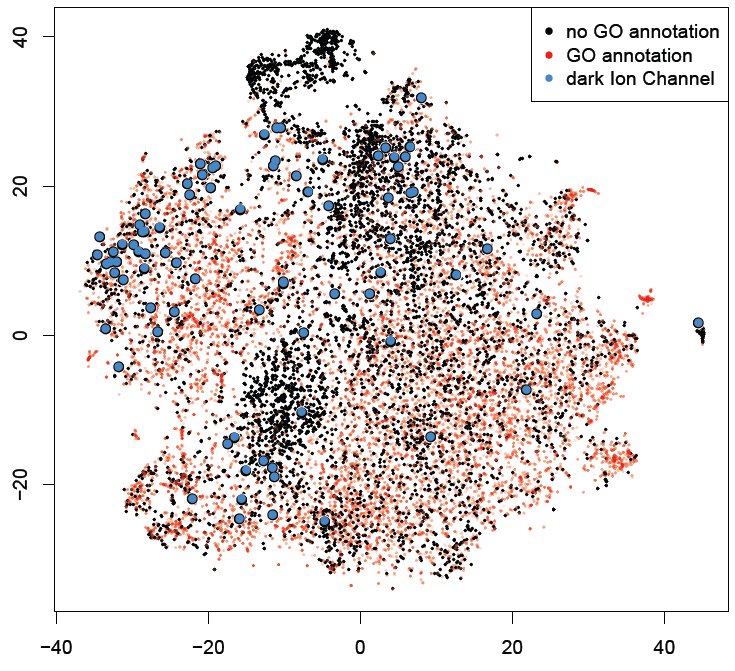
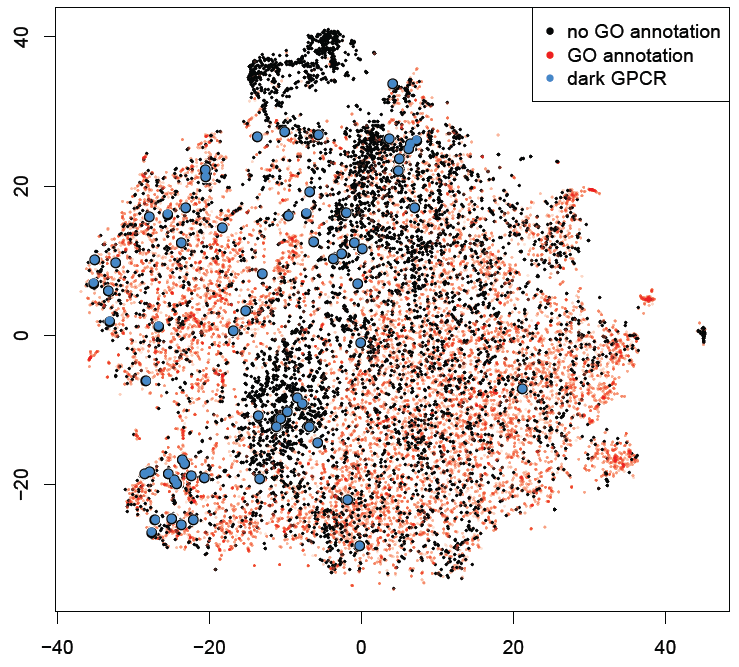
Figure 1 a) To show the correctness of term ranking, area under the curve (AUC) from the ROC was calculated for all genes with existing annotations in GO. For GO biological processes, the average AUC for retrieving already annotated terms is 0.765. b-d) t-SNE plots of all human genes based on their co-expression similarity, where each gene is a point on the plot. Genes are colored based on the number of GO terms associated with each genes. Black dots are genes with no GO terms. Blue dots highlight the under-studied kinases, GPCRs and ion channels.
Examples of GO annotations for the three under-studied kinase: PKMYT1, NEK4, PKN3; and the GPCR: ADGRD2
| PKMYT1 This gene encodes a member of the serine/threonine protein kinase family. The encoded protein is a membrane-associated kinase that was reported to negatively regulates the G2/M transition of the cell cycle by phosphorylating and inactivating cyclin-dependent kinase 1. The activity of the encoded protein is regulated by polo-like kinase 1. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. This kinase is listed as an under-studied kinase by IDG, and here we show the results of predicting GO biological processes terms for PKMYT1. |
| NEK4
NEK4 (NIMA Related Kinase 4) is a kinase known to be associated with Meckel Syndrome 1. It was reported that this kinase plays a role in sweet taste signaling. GO annotations related to this gene are few and include tyrosine kinase activity. This kinase is listed as an under-studied kinase by IDG, and here we show the results of predicting GO biological processes terms for NEK4. |
| PKN3
PKN3 (Protein Kinase N3) was reported to be a member of the Salmonella infection pathway (KEGG). GO annotations related to this gene include protein tyrosine kinase activity. This kinase is listed as an under-studied kinase by IDG, and here we show the results of predicting GO biological processes terms for PKN3. |
| ADGRD2
ADGRD2 (Adhesion G Protein-Coupled Receptor D2) is an under-studied G-protein coupled receptor. An important paralog of this gene is ADGRL1. This kinase is listed as an under-studied kinase by IDG, and here we show the results of predicting GO biological processes terms for ADGRD2. |