About
ARCHS4: Massive Mining of Publicly Available RNA-seq Data from Human and Mouse
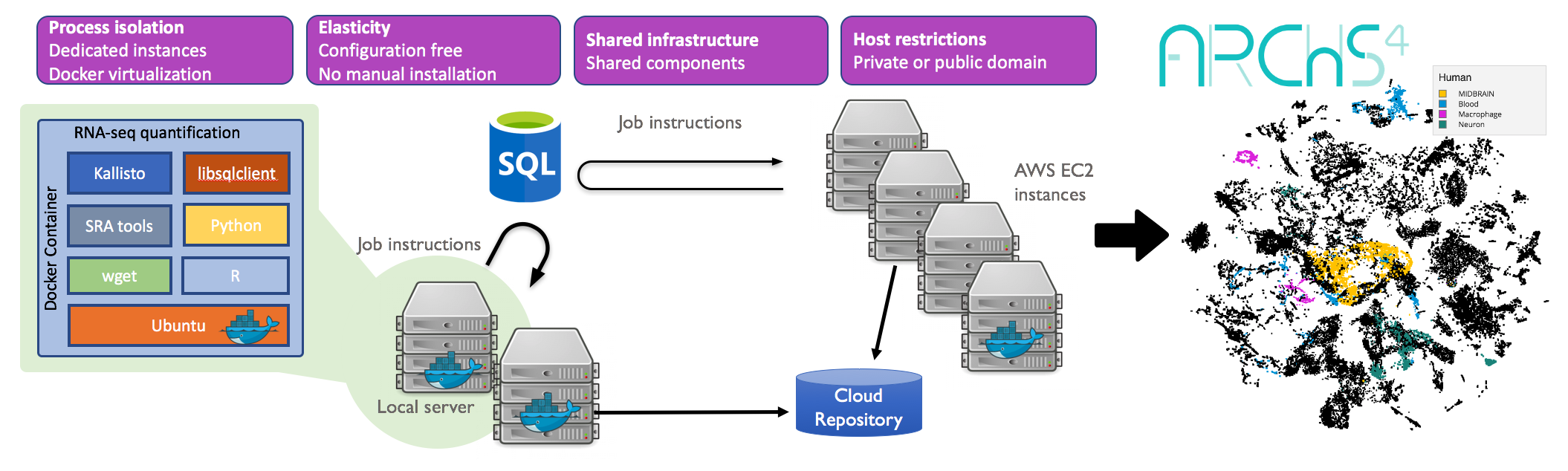
All RNA-seq and ChIP-seq sample and signature search (ARCHS4) is a resource that provides access to gene and transcript counts uniformly processed from all human and mouse RNA-seq experiments from the Gene Expression Omnibus (GEO) and the Sequence Read Archive (SRA).
The ARCHS4 website provides the uniformly processed data for download and programmatic access in H5 format, and as a 3-dimensional interactive viewer and search engine.
Users can search and browse the data by metadata enhanced annotations, and can submit their own gene sets for search. Subsets of selected samples can be downloaded as a tab delimited text file that is ready for loading into the R programming environment. To generate the ARCHS4 resource, the kallisto aligner is applied in an efficient parallelized cloud infrastructure. Human and mouse samples are aligned against GRCh38 and GRCm39 with Ensembl annotation (Ensembl 107).
ARCHS4 Workshop
The workshop will be presented by Alexander Lachmann, Ph.D., who is an assistant professor at Ma’ayan laboratory and developed ARCHS4.
During the workshop, we will cover
- ARCHS4
- Introduction
- Programmatic approach
- Use cases
- Live Demo
- Kallisto, one of the leading algorithms on gene alignment
Ma’ayan Lab
The Ma’ayan Laboratory develops computational and mathematical methods to study the complexity of regulatory networks in mammalian cells. We apply machine learning and other statistical mining techniques to study how intracellular regulatory systems function as networks to control cellular processes such as differentiation, dedifferentiation, apoptosis and proliferation. We develop software systems to help experimental biologists form novel hypotheses from high-throughput data, while aiming to better understand the structure and function of regulatory networks in mammalian cellular and multi-cellular systems.
Feel free to contact us at maayanlabapps@gmail.com