How To Use
DrugBot Functions
Drug Summary
DrugBot enables users to submit a drug or small molecule of interest and outputs information pertaining to the term searched. The /drug command provides a summary about the drug and links to various databases.
The drug is searched through DrugBank and L1000FWD metadata. DrugBot will output links to drug landing pages for the drug based on which of these databases it is found in.
✦ Input Format: /drug <drug name>
✦ Example:
- Input:
/drug imatinib - Output:
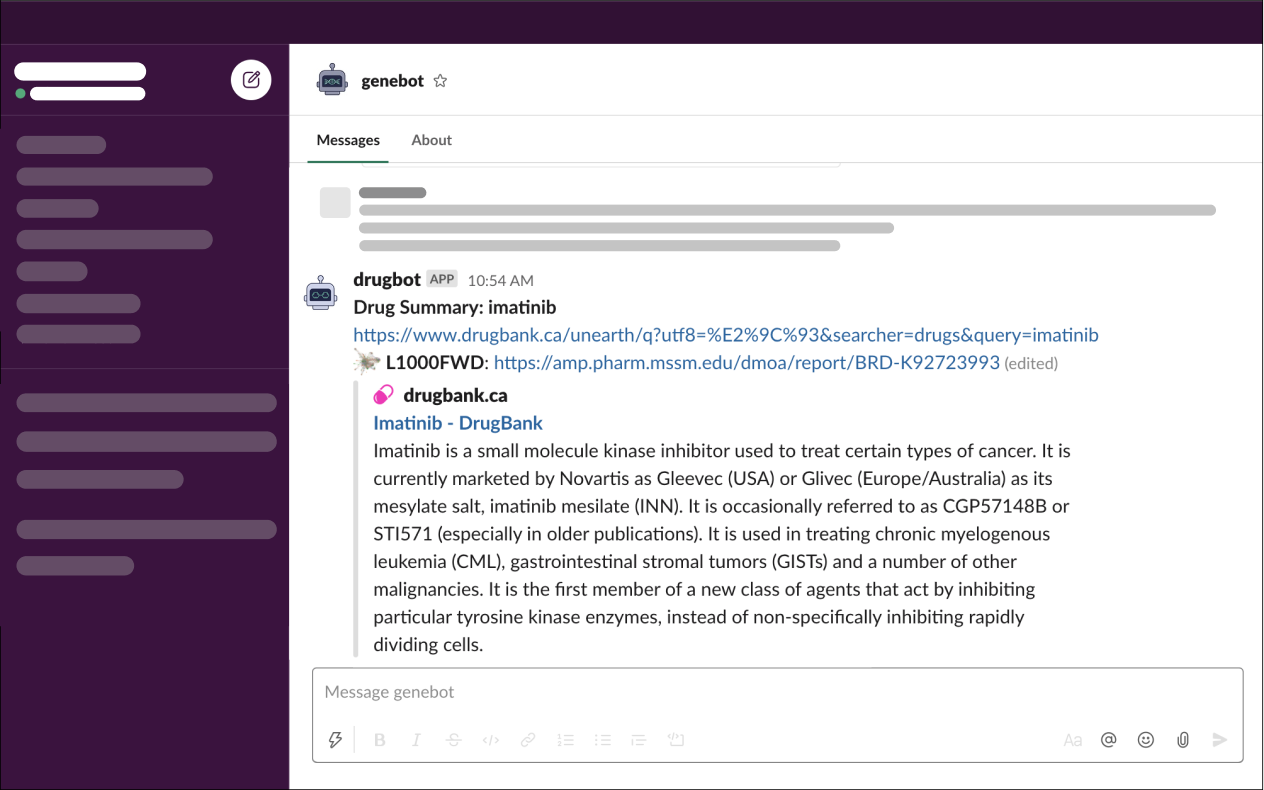
Drug Enrichment Analysis
DrugBot enables users to submit a drug set for enrichment analysis. The /drugset command can be used to perform drug enrichment analysis by listing valid drug names following the command. Alternatively, users can upload a CSV file with the drug set for analysis by calling @drugbot followed by drugset. Additionally, users can specify which drug-set library they want to be displayed in their results in Slack.
The drug set is analyzed using the DrugEnrichr API. The bot will output a link to DrugEnrichr for the drug set entered as well as summary statistics about the drug set. A bar graph displaying the top 10 enriched terms for the user selected drug-set library will be attached as a pdf file.
✦ Input Format:
- Input (no library specified):
/drugset drug1, drug2, drug3...
- Input (library specified):
/drugset [library, drug1, drug2, drug3...]- Drug list can be space or comma separated.
- Library specified input must be contained by brackets.
- Input (file upload):
@drugbot set *file*@drugbot set library *file*- Here is a sample file that is properly formatted with a drug set.
✦ Example:
- Input:
@drugbot set DB *file* - Output:
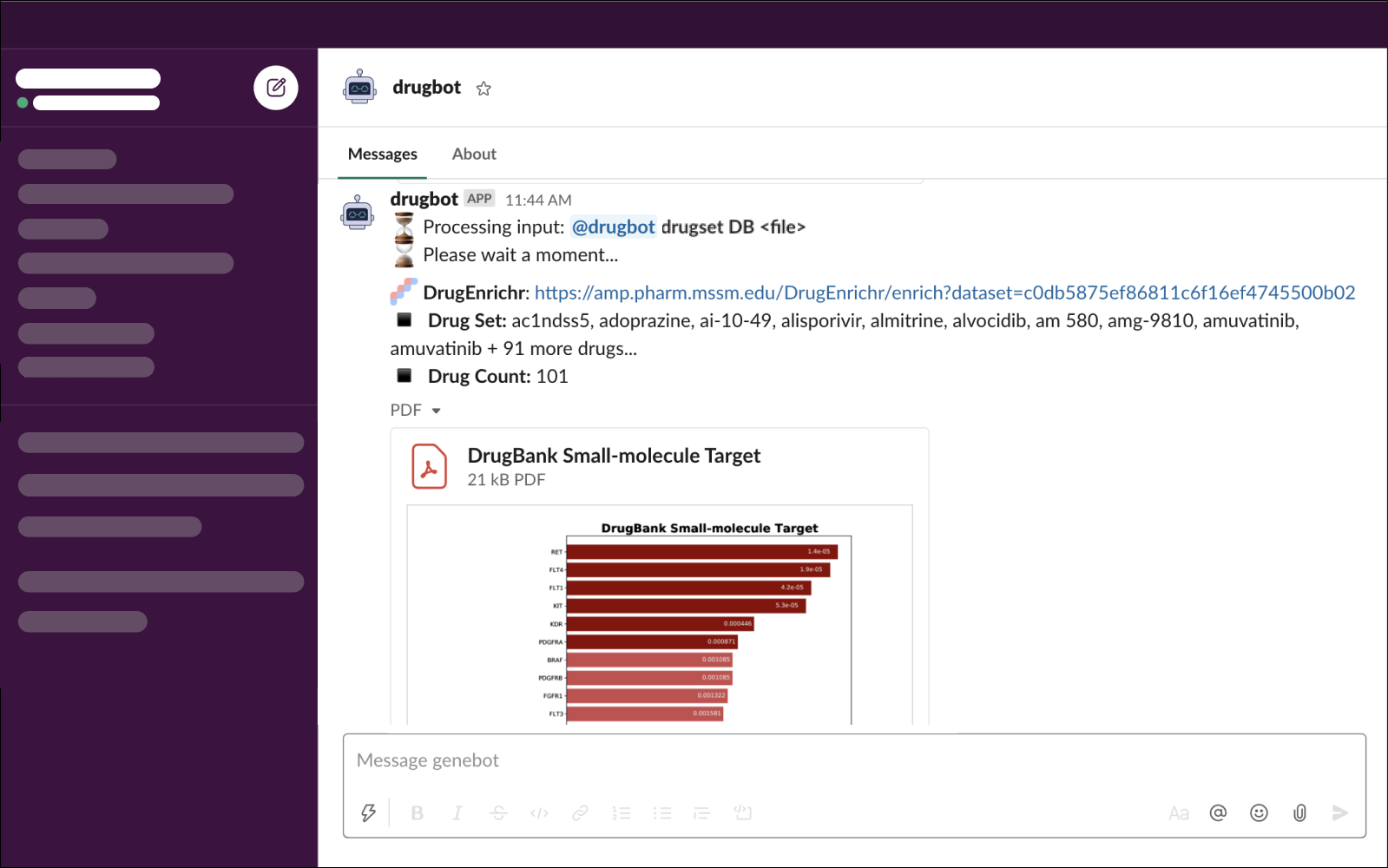
✦ Drug Libraries: When specifying a library for output, DrugBot will recognize full DrugEnrichr library names or predefined short hands. Full DrugEnrichr library names can be found here. Library short hands are available for several widely-used drug set libraries.
- Library short hands include:
- GSA = Geneshot_Associated
- GSPE = Geneshot_Predicted_Enrichr
- DB = DrugBank_Small-molecule_Target
- L1000D = L1000FWD_GO_Biological_Processes_Down
- STITCH = STITCH_Target-seq_2015
- SIDER = SIDER_Side_Effects
✦ Help Commands:
There are several built in help commands for performing analysis using DrugBot that can be called in Slack to provide concise summaries about different app features and functions. A list of all commands can be found by calling: @DrugBot help
/drug-help→ provides a quick guide on using DrugBot to get information about a drug/drugset-help→ provides summary of how to use DrugBot for enrichment analysis/drugset-help library?→ provides instructions on how to specify library for output/drugset-help fileupload?→ provides instructions on how to upload file for analysis/drugset-help slashcommand?→ provides instructions on how to navigate /drugset command
If you have any questions about how to use DrugBot, please contact us at maayanlabapps@gmail.com.
 DrugBot
DrugBot